在江苏省中医药管理局科研项目(批准号:YB2020088)、扬州市“绿扬金凤”卫生创新领军人才基金项目(批准号: YZLYJF2020WSCX037)等资助下,南京鼓楼医院集团仪征医院王小红主任医师团队在红景天苷治疗重症急性胰腺炎机制方面取得进展。研究成果以“红景天苷通过调节 miR-217-5p/YAF2 轴减轻重症急性胰腺炎触发的胰腺损伤和炎症(Salidroside alleviates severe acute pancreatitis- triggered pancreatic injury and inflammation by regulating miR-217-5p/YAF2 axis)”为题,于2022年8月10日在线发表于《国际免疫药理学》(International Immuno- pharmacology)。文章链接:https://doi.org/10.1016/j.intimp.2022.109123

急性胰腺炎(Acute pancreatitis)是临床常见的急腹症之一,其以轻症多见,病程呈自限性,其中10%~30%的患者会进展为重症急性胰腺炎(Severe acute pancreatitis, SAP)。SAP病情进展快,病情危重,常并发多脏器功能不全综合征,病死率高达20%~30%,给家庭和社会带来沉重负担。因此,探索SAP的发病机制,寻找有效的治疗药物及方法显得尤为重要。
该团队通过鉴定与红景天苷相关的miRNA并探索了其潜在机制。该团队通过建立SAP大鼠动物模型和miRNA微阵列,确定红景天苷对miRNA表达谱的影响,并通过实时定量PCR验证它们的变化。该团队观察到,在这些miRNA中,经红景天苷处理后miR-217-5p显著下调。miR-217-5p过表达可以通过直接靶向 YY1相关因子2 (YAF2)的3'UTR来逆转由红景天苷介导抑制的免疫反应。
该研究揭示了红景天苷通过调节炎症反应发挥对SAP的保护作用,而在调节炎症反应的机制中涉及了miR-217-5p。此外,抑制miR-217-5p可通过靶向YAF2的3'UTR 促进YAF2表达,随后激活p38 MAPK 通路。(见Figure1, 2, 3, 4)
该研究结果为红景天苷治疗SAP的作用机制提出了新的见解,并有助于制定SAP的有效治疗策略。
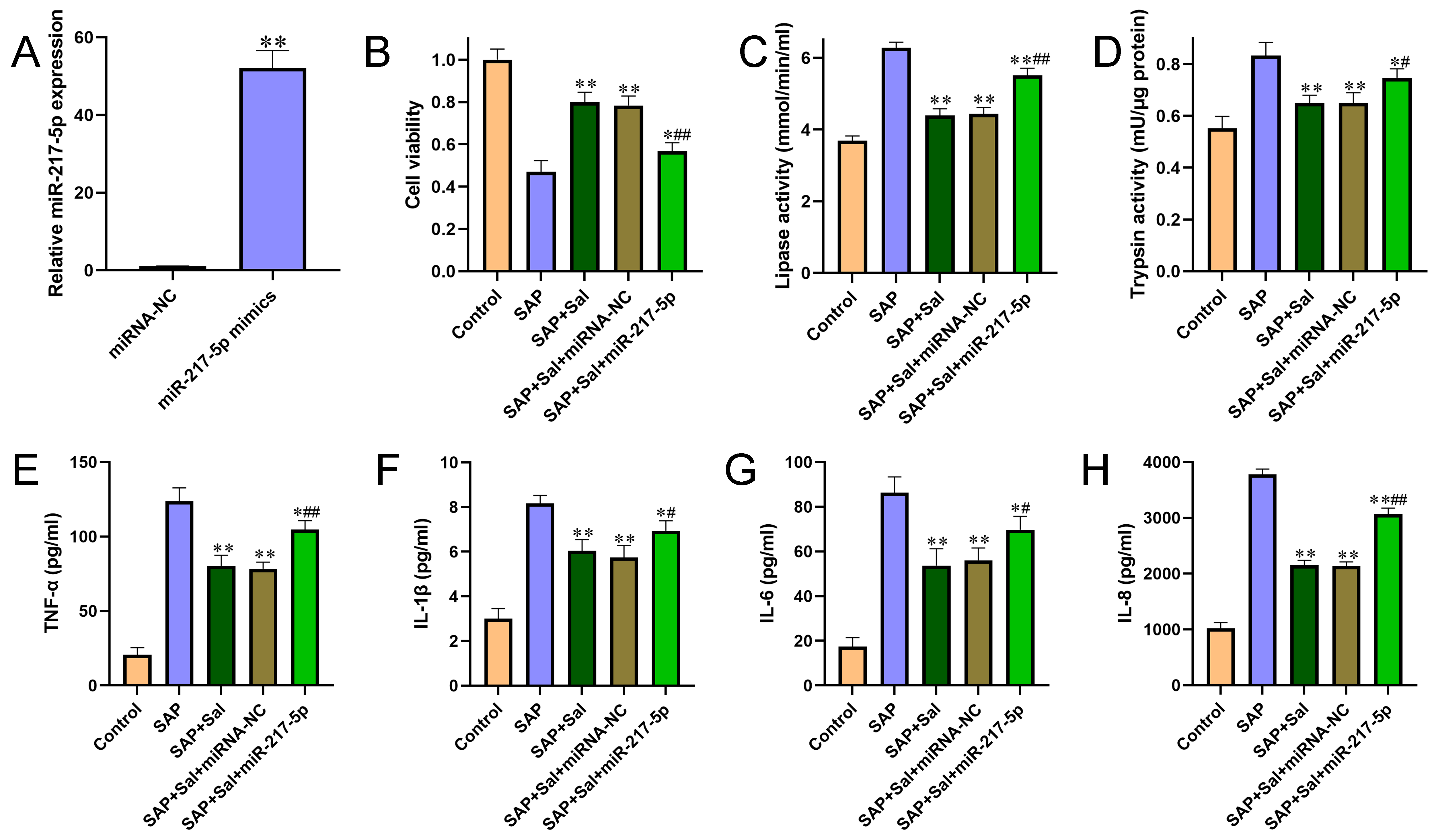
Figure 1 Overexpression of miR-217-5p aggravated cell injury and promoted pro-inflammatory cytokine production. (A) The overexpression efficiency of miR-217-5p mimics was confirmed by qRT-PCR. (B) The effect of miR-217-5p overexpression on cell viability was evaluated by CCK-8 assay. (C-D) The effects of miR-217-5p overexpression on activities of amylase (C) and lipase (D) were detected by automatic biochemical analysis. (E-H) The levels of pro-inflammatory cytokines TNF-α (E), IL? 1β (F), IL-6 (G) and IL-8 (H) from cultured cell medium were determined by ELISA kits according to instructions of the manufacturer. (I-K) Pearson’s correlation analysis was performed to analyze the association between miR-217-5p expression and pancreatic injury score (I), serum amylase activities (J) and serum lipase activities (K). For B-H, *P<0.05, **P<0.01 compared with SAP group, #P<0.05, ##P<0.01 compared with miRNA negative control group.
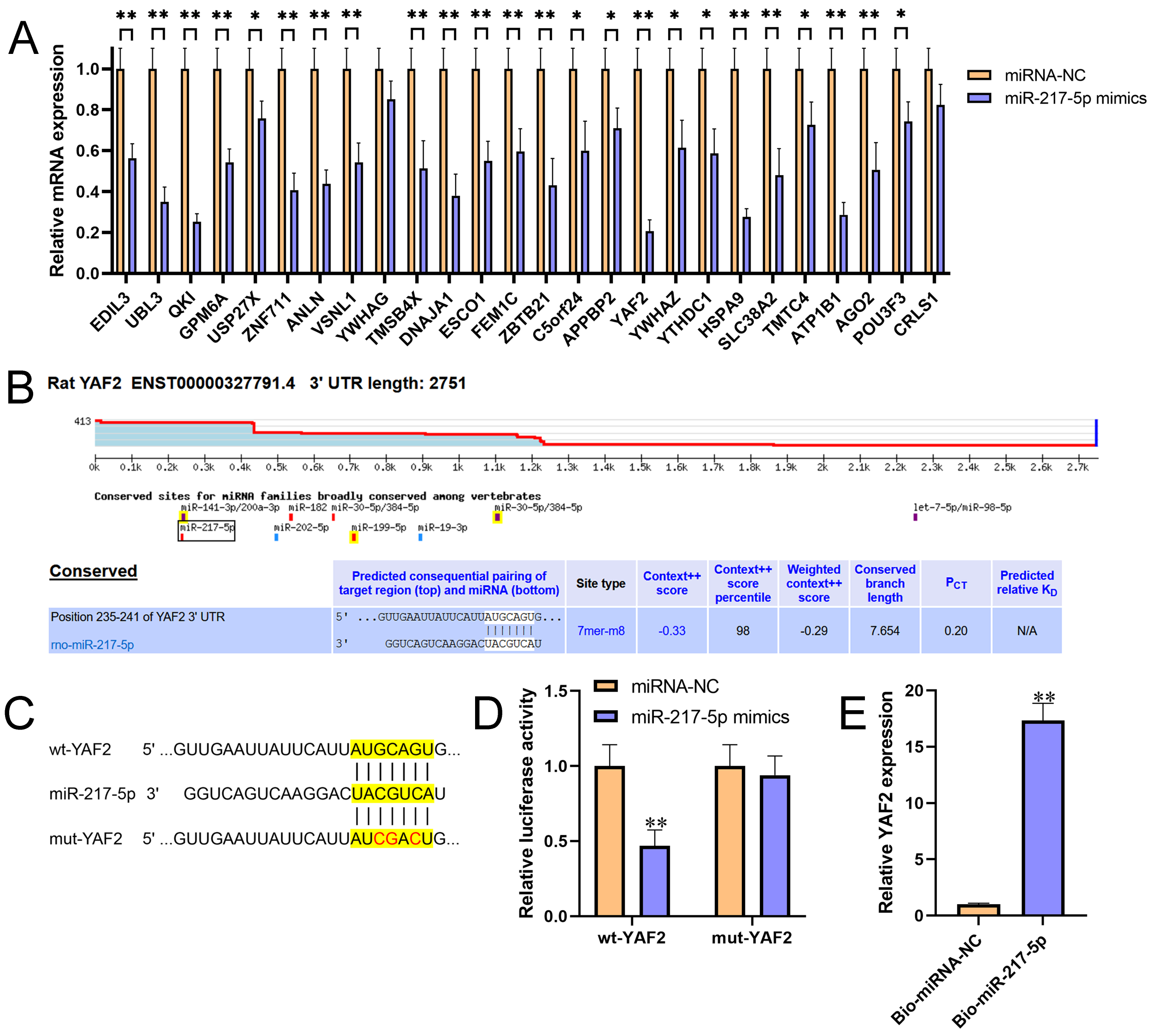
Figure 2 miR-217-5p directly targeted the 3’UTR of YAF2. (A) Relative expression levels of 26 candidate target genes for miR-217-5p were detected by qRT-PCR in AR42J cells after transfected with miR-217-5p mimics or miRNA negative control. (B) The binding position and complementary sequences of miR-217-5p in the 3’UTR of YAF2, which was predicted by TargetScan. (C) The wt- and mut-YAF2 3’UTR sequences for miR-217-5p. (D) Luciferase reporter constructs containing wt- and mut-YAF2 3’UTR were co-transfected with miR-217-5p mimics or miRNA negative control into AR42J cells. After 48h transfection, the luciferase activities were measured. (E) Relative level of YAF2 recruited by Bio-miR-217-5p or Bio-miRNA-NC probes were measured by miRNA pull-down assay. **P<0.01.
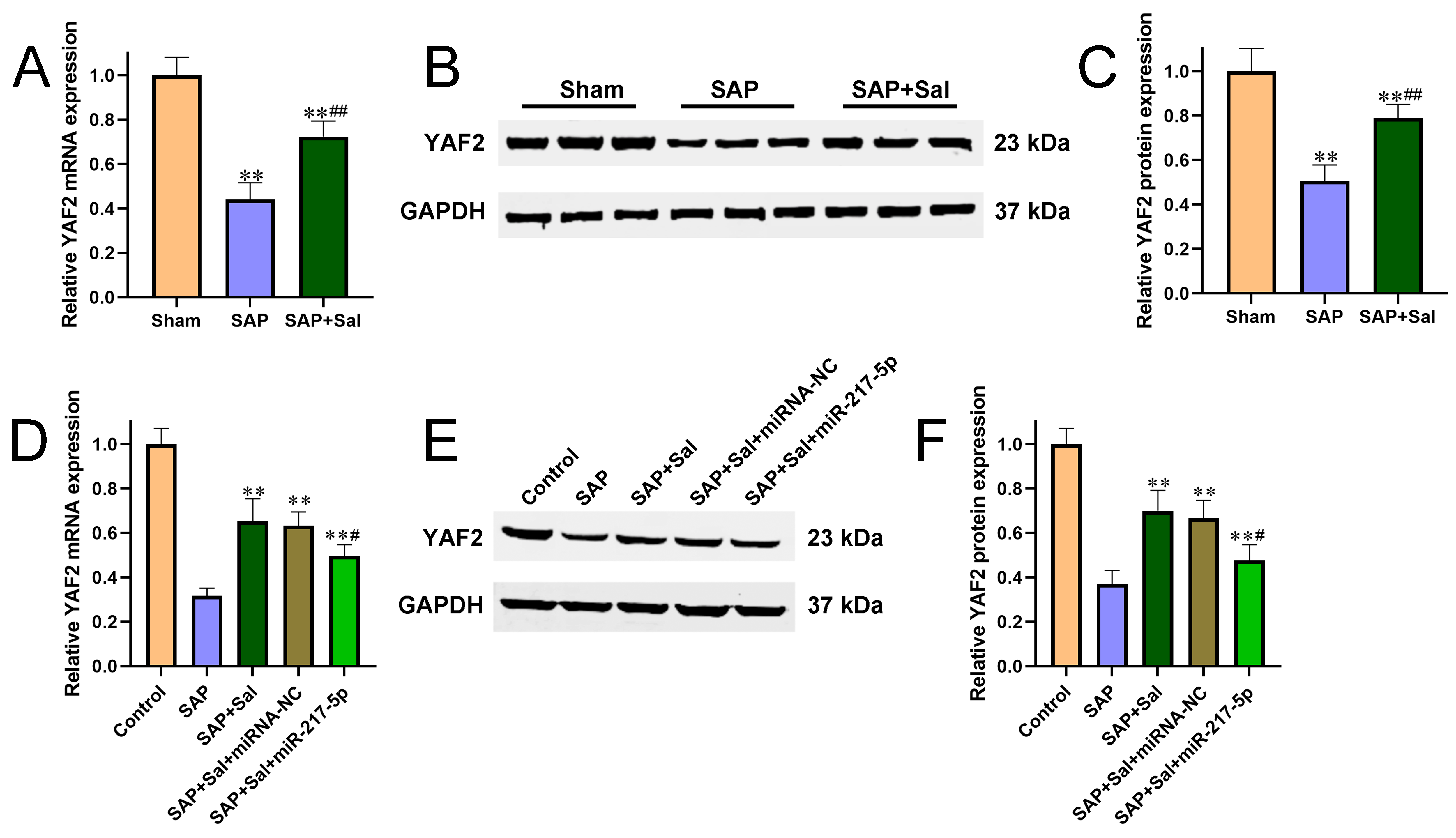
Figure 3 Sal upregulated YAF2 expression via downregulating miR-217-5p. (A-C) The mRNA (A) and protein (B-C) levels of YAF2 in SAP rat pancreatic tissues were determined by qRT-PCR and Western blot, respectively. (D-F) The mRNA (D) and protein (E-F) levels of YAF2 in AR42J cells were determined by qRT-PCR and Western blot, respectively. **P<0.01 compared with sham group, #P<0.05, ##P<0.01 compared with SAP group.
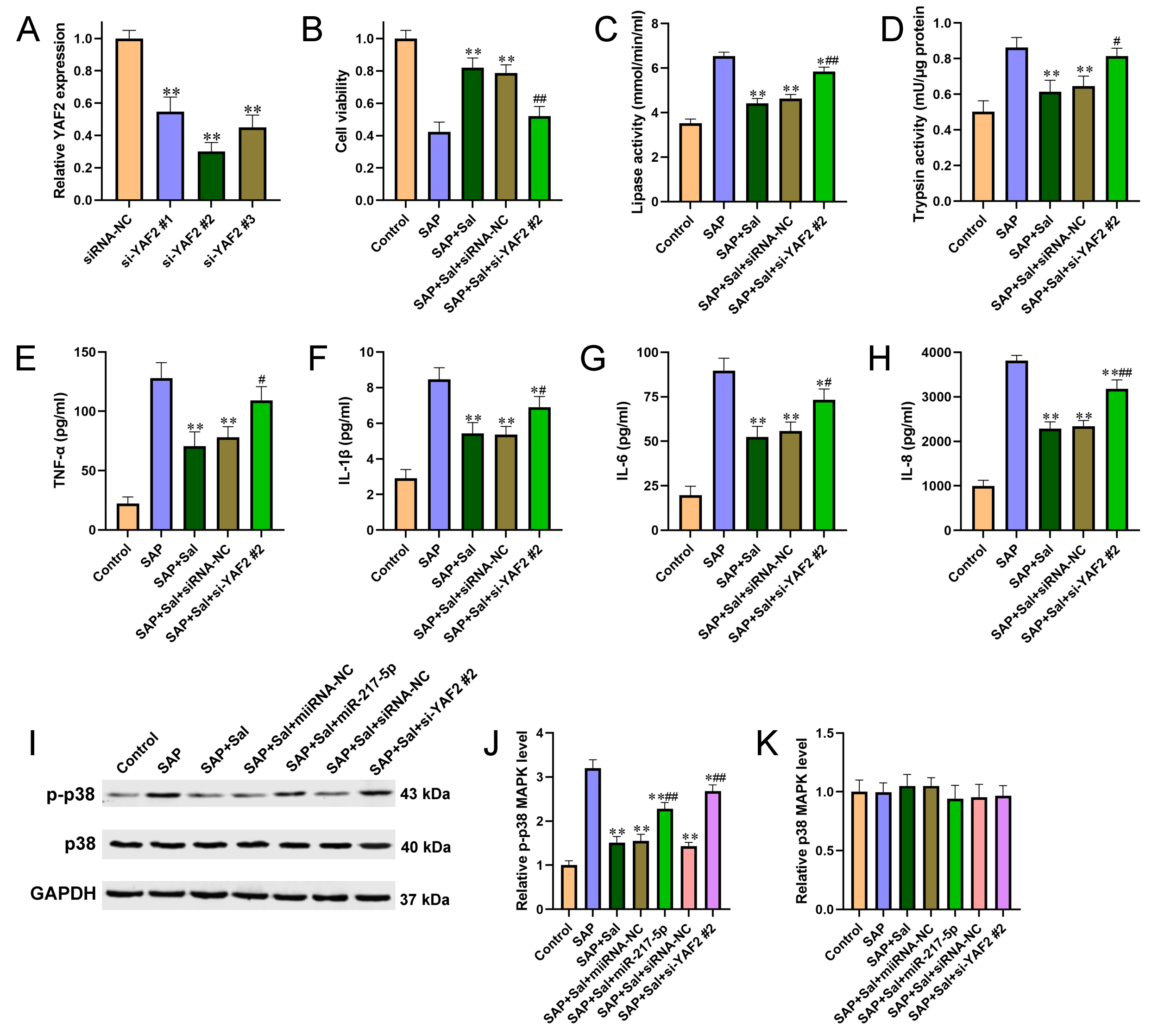
Figure 4 Knockdown of YAF2 reversed Sal induced alleviation of cell injury and inflammation in a YAF2-dependent manner and involving in p38 MAPK pathway. (A) siRNA specific for YAF2 was transfected into AR42J cells and the knockdown efficiency was validated by qRT-PCR. (B) Cell viability was detected by CCK-8 assay. (C-D) The effects of YAF2 knockdown on activities of amylase (C) and lipase (D) were determined by automatic biochemical analysis. (E-H) The effects of YAF2 knockdown on the pro-inflammatory cytokines TNF-α (E), IL? 1β (F), IL-6 (G) and IL-8 (H) were determined by ELISA kits. (I) Western blot was performed to detect the expression levels of p38 and p-p38 in AR42J cells after different treatment. (J-K) Quantitative analysis of p-p38 (J) and p38 (K) from 7I. For A, **P<0.01 compared with siRNA negative control. For B-K, *P<0.05, **P<0.01 compared with SAP group, #P<0.05, ##P<0.01 compared with miRNA or siRNA negative control group.